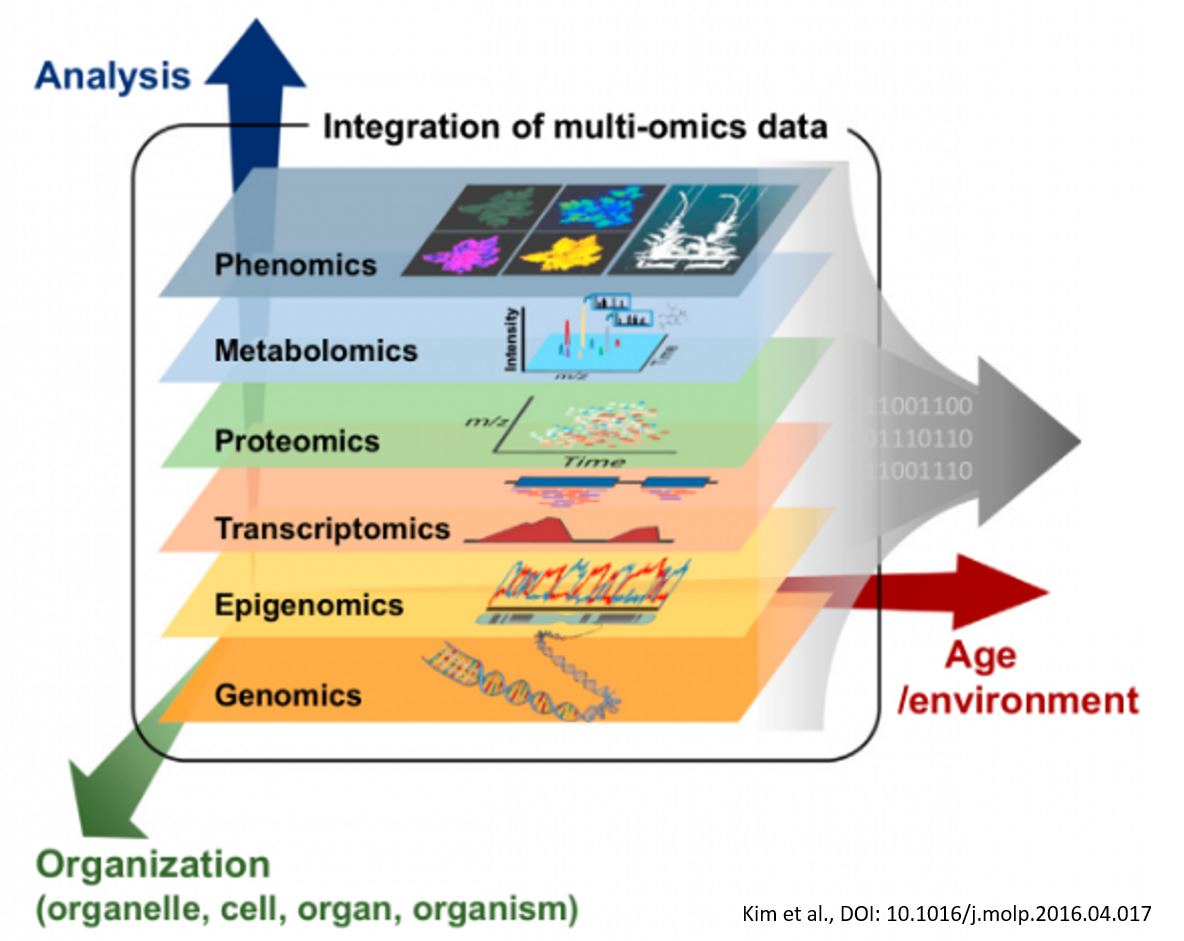
Multi-omics
In molecular biology, the flow of genetic information is classically described by the Central Dogma, stating that genetic code is transcribed into RNA, which is in turn translated into proteins. These are the functional units which maintain cellular homeostasis and the source of cell type specific functions. However, recent developments in the fields of epigenetics and metabolomics have significantly challenged the Central Dogma. The genetic architecture was shown to directly affect genetic activity while the metabolism, previously thought of as one of the final outputs of the genome, was found to be a major source of input and genetic regulation by acting as an environmental sensor. The omics group of our lab integrates state-of-the-art transcriptomics and metabolomics assays with epigenomics to characterise cellular activity in bone tissue engineering applications, cancer and lymphangiogenesis. Simultaneously, our computational team is tackling the rising challenge imposed by the scale of these datasets. Together, we aim to gain an integrated understanding of life’s most fundamental processes.
Currently in this project: Tim, Marc, Bernard, Liesbeth, Gabriele
