In silico systems biology
In silico systems biology aims at using general mathematical and computational tools to get insights into biological and medical questions, mainly through the study of regulatory networks.
Different modeling strategies are of concern belonging to causal (mechanistic) or correlation (data driven) families. A few examples of interest in our group are: ordinary and partial differential equations, logical modeling, additive models, agent based models, stochastic and data-driven modeling.
Currently in this project: Raphaëlle, Sophie, Marc, Tim, Bernard, Alessio, Luiz, Loïc
–Raphaëlle
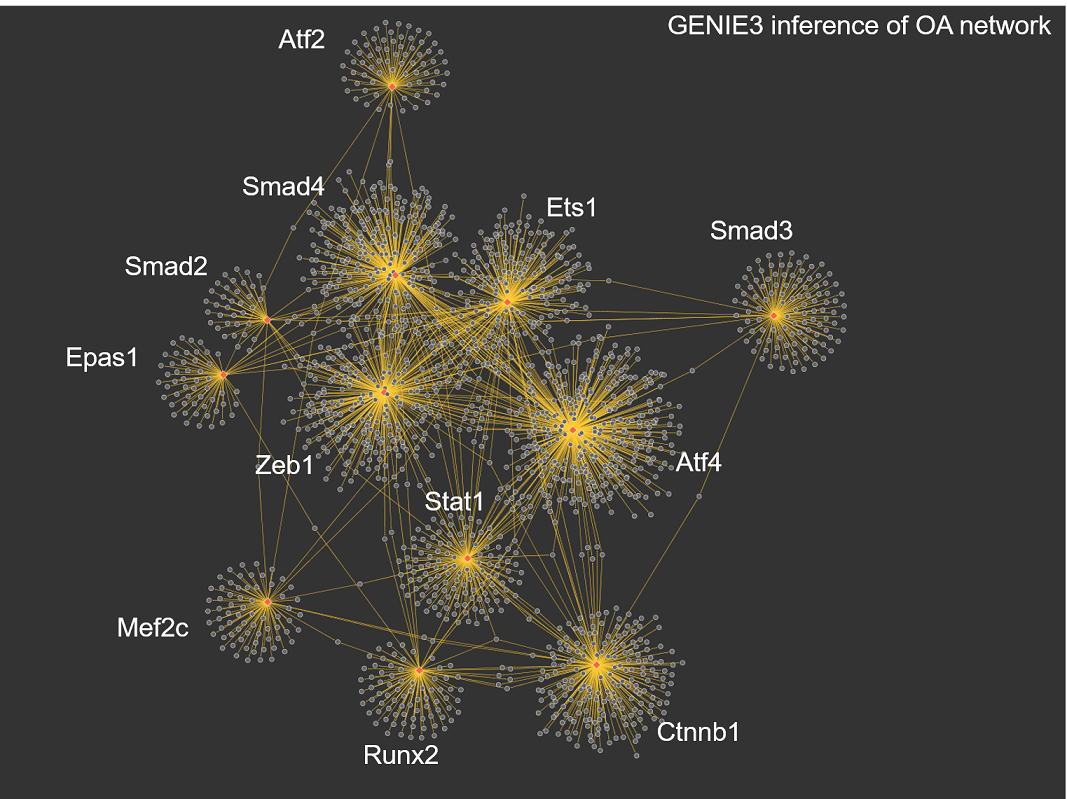
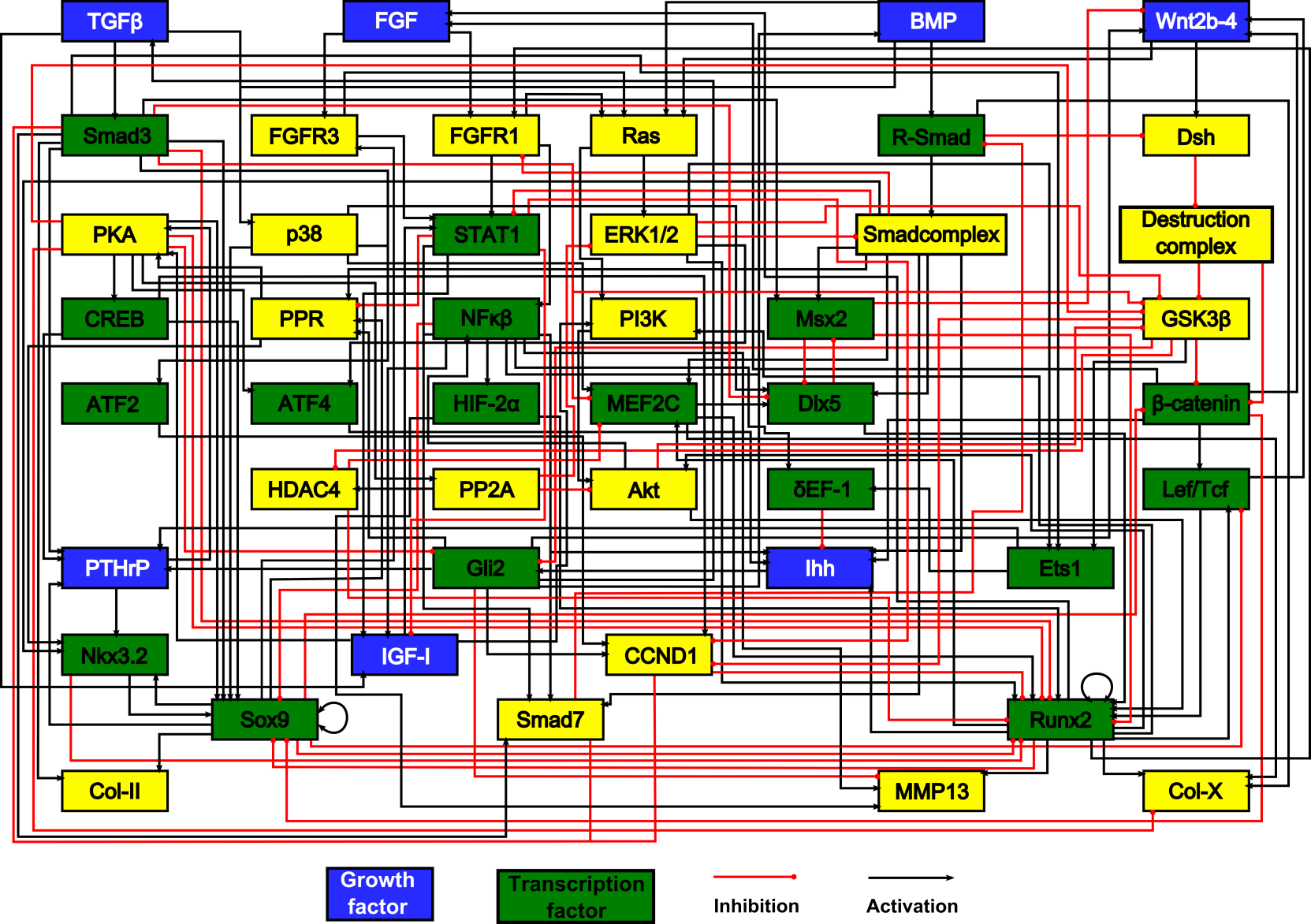
We combine mechanistic modeling and data-driven approaches in order to study chondrocyte differentiation in the context of bone Tissue engineering (TE) and Osteoarthritis (OA). We carry out mechanistic modeling using additive qualitative formalism mainly, whereas we use machine-learning methods to infer regulatory network based on gene expression data.
–Morgan
We investigate the hypertrophic switch in human skeletal progenitors through the use of deterministic models, mainly using ordinary differential equations (ODEs). In particular, we study the irreversible bistable behavior of this switch, the interactions between the BMP and Wnt pathways and the receptors-ligand binding dynamics of the BMP pathway.
–Sophie
We study the process of lymphangiogenesis with mathematical and computational tools. We will address the questions behind this process by the development of in silico models, using both open-ended (RNA-Seq and bioinformatics analysis) and hypothesis-driven approaches (specific set-ups and mechanistic models), which together will provide information and insights that are currently lacking. In silico models at different spatiotemporal scales and levels of complexity will be considered.
–Marc
We extend a stochastic model of protein synthesis to unravel regulation upper layers affecting protein expression above the transcriptional level. We combine a data driven systematic mining approach and a mechanistic top-bottom approach to understand Ribo-seq experiments results and t-RNA reprogramming that selectively express transcripts with specific codon usage biases. The first proof of concept of our computational modeling addresses targeted therapies in melanomas.
