Combining insight and expertise from biomedical sciences and engineering sciences, the Tissue Engineering (TE) group of our lab aims to develop strategies to develop living implants for large osteochondral defects. We are continuously developing new tools and technologies to assist the translation of the lab-based solutions to the clinic. The majority of these tools are computer models (in silico tools) of all processes involved in the TE pipeline, from network models for cell differentiation to the prediction of in vivo responses to our treatments. The in silico models use a variety of model technologies and serve to increase fundamental understanding as well as to perform in silico screening before progressing to the in vitro/in vivo stage. In silico models of biomaterial-induced biology in combination with our in-house3D (bio)printing facilities allow to design, produce and test new scaffolds for a variety of applications. An in-house developed perfusion bioreactor together with its digital twin allows to test both fundamental biological questions regarding the influence of the local cell environment and translational engineering questions regarding process control and optimisation.
Tissue Engineering

Systems Biology
Omics: In molecular biology, the flow of genetic information is classically described by the Central Dogma, stating that genetic code is transcribed into RNA, which is in turn translated into proteins. These are the functional units which maintain cellular homeostasis and the source of cell type specific functions. However, recent developments in the fields of epigenetics and metabolomics have significantly challenged the Central Dogma. The genetic architecture was shown to directly affect genetic activity while the metabolism, previously thought of as one of the final outputs of the genome, was found to be a major source of input and genetic regulation by acting as an environmental sensor.
Systems biology: The systems biology group of our lab integrates state-of-the-art transcriptomics and metabolomics assays with epigenomics and mechanistic systems biology approaches to characterise cellular activity in bone tissue engineering applications, cancer and lymphangiogenesis. Simultaneously, our computational team is tackling the rising challenge imposed by the scale of these datasets and the complexity of interconnected networks. Together, we aim to gain an integrated understanding of life’s most fundamental processes.
Computational Biomechanics
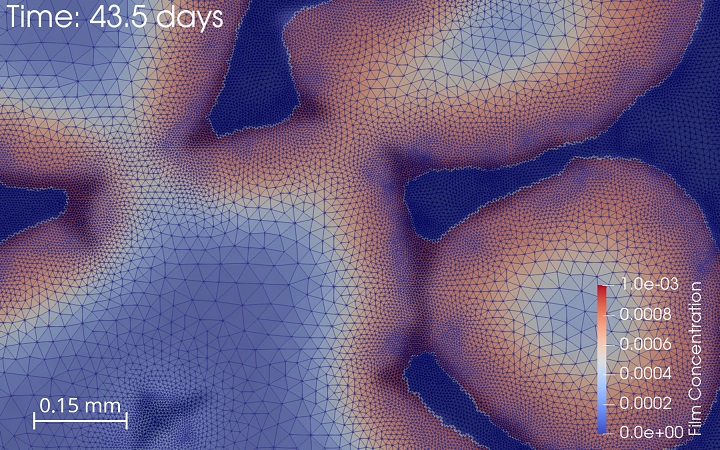
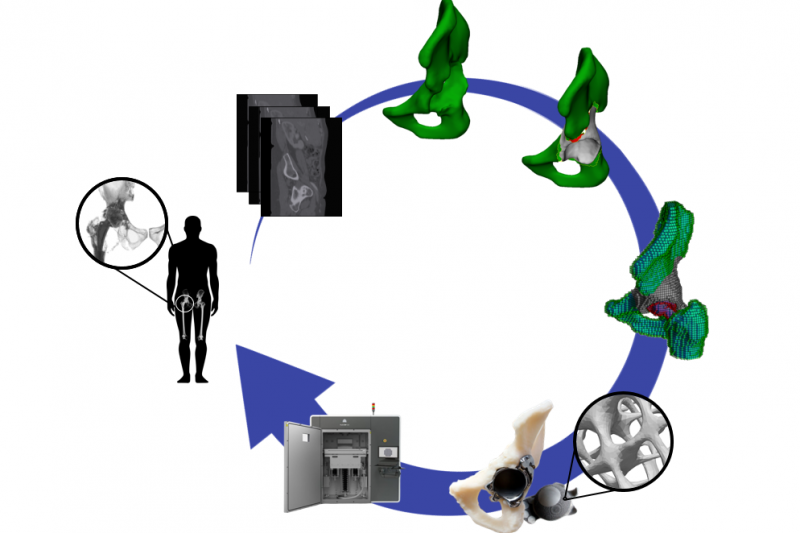
The interaction between mechanics and biology is at the heart of many physiological and pathophysiological processes in the skeleton. The computational biomechanics group of our lab uses computational techniques (finite element modeling) to characterise the mechanical environment of joints, developing limbs and experimental set-ups. We use this quantification, in combination with computer models of the biological response, to optimize the design of hip and knee implants and to optimize the design of in vitro bioreactor set-ups that aim to recreate relevant in vivo conditions. We furthermore use these tools to gain insight into the biological processes of limb development and bone regeneration.
Biophysics
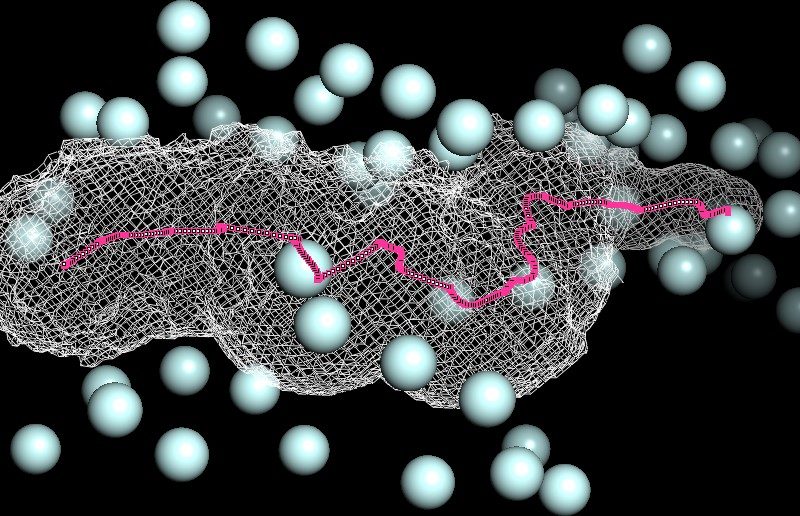
Our team set out to develop models of life processes such as mRNA translation. We explore the biophysics of molecular machines like ribosomes and investigate, for instance, how electrostatics forces at the molecular level impact on protein synthesis rate. The publicly available solved structures of intracellular biological components by X-ray crystallography or cryo-electron microscopy are valuable resources helping us in the design or parametrization of our biophysical models. The aim is to comprehend how fundamental laws of physics and chemistry constrain biological phenomena.
Figure: 3D map of phosphorus atoms (light blue) in the negatively charged phosphates moieties around the ribosome exit tunnel (white mesh) ; 93 Angstroem length centro-axial line of the ribosomal tunnel (magenta). Ribosome large subunit X-ray solved structure (PDB codename ‘4V9F’) of Haloarcula marismortui.
Our Research Team

Liesbet Geris

Tim Herpelinck

Sophie Bekisz

Fernando Pérez Boerema

Marc Joiret

Laura Lafuente-Gracia

Bernard Staumont

Gabriele Nasello

Ayse Kose

Edoardo Borgiani

Ahmad Alminnawi

Luiz Ladeira

Liesbeth Ory

Margaryta Ivanets

Pieter Ansoms

Loïc Comeliau

Tom Verbraeken

Sophie Nguyen

Oriana De Becker












